Last year I collaborated with cross-institution researchers from Bangladesh led by BCSIR on the severe Dengue outbreak in 2021. The study has been published in the BMC Virology Journal recently.
Dengue remains a dangerous endemic disease causing many deaths and suffering in Bangladesh.
My contribution was to analyze the phylodynamics of the Dengue virus (DENV). The DENV3 serotype dominated the 2021 outbreak. We confirmed a previously reported genotype shift (DENV3-II to DENV3-I).
Right after the 2006–2009 DENV3 outbreak, the genotype shift presumably happened in Bangladesh (tMRCA from genotype I DENV3 sequences from Bangladesh was 2013).
Another interesting fact. The 2021 epidemic isolates, which share strong similarities, are well separated from the 2017 epidemic isolates in Bangladesh but interleaved by two DENV3 isolates sampled in 2019. Interestingly, these two isolates, 19XN13542 and 19XN14065, were sampled in China from travelers returning from Bangladesh.
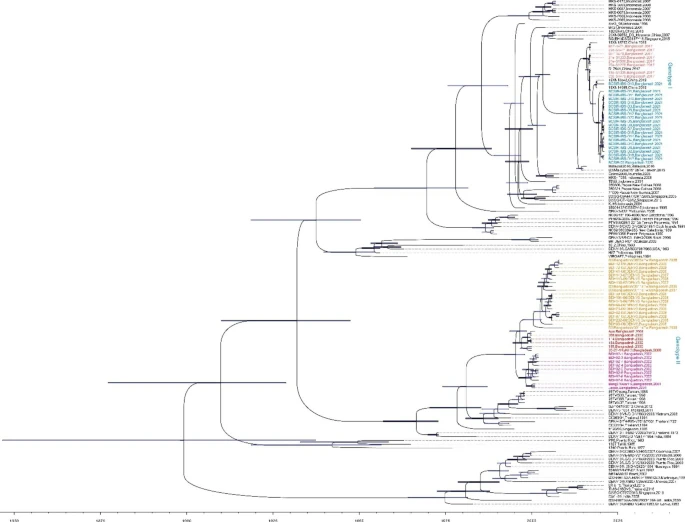
Figure: Phylodynamic analysis showed a clad shift in the DENV3 in Bangladesh. Source: Original paper.
So what is going on? A transboundary movement of the virus? Only further research can tell. But this highlights the importance of genomic surveillance for these infectious diseases.
This year’s cases are showing a high prevalence of DENV3, too (>60%) followed by the other strains, according to IEDCR.
Check out the article for detail:

Leave a Reply