After completing the human genome sequencing in the 2000s, much of our DNA, even more than half, was considered unnecessary. It was called “junk DNA,” “broken genes” trapped in the genome’s prison, and the DNA fossils of viruses that have been silenced. It was thought these destroyed DNA elements had no purpose and were unrelated to changes.
However, in the past decade, research has shown that some of this “junk DNA” is not entirely useless. Compared to the entire genome, the amount of functional genes is minimal, only 2 percent, which can produce various proteins. It is now known that certain so-called “non-coding” DNA helps regulate the expression of different genes. However, whether these DNA controllers are essential or potentially harmful to the body is still debated among scientists.
In 2013, research from the ENCODE project revealed that non-coding DNA, which was previously considered junk, actually performs important functions, leading to controversy and discussions. Since then, there has been further research on non-coding DNA. In this article, we will learn about some of the new research that does not support the concept of “junk DNA.”
Junk DNA can be essential in mammalian development
Researchers from the University of California, Berkeley, and Washington University have worked on the transposons. Transposons involves the selfish DNA elements that integrate into various parts of the genome. A specific family of transposons is derived from the DNA of different ancient viruses. When these viruses infect a host, their DNA elements become integrated into the genome of certain individuals. During the long evolution of the host’s genome and the viral genome, these “foreign” elements underwent evolution as transposons. This research suggests that transposons from viruses are inevitable for the survival of certain hosts. When researchers removed these transpositions from the genomes of mice, more than half of them died before birth.
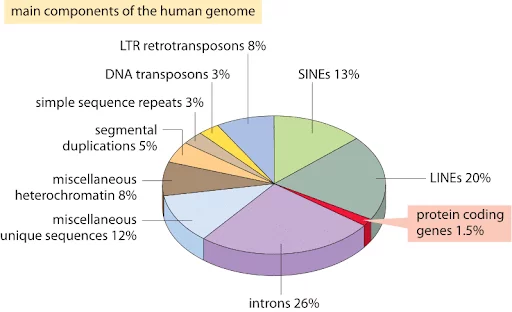
Figure: Only 1.5-2% of human genome are protein coding genes. So what are the rest?
The presence of “junk” DNA can also be relevant to lactating animals, as observed in this research for the first time. Lin He, one of the scientists involved, explains that at different stages of embryonic development in mice, the timing of expansion in various cells and the mother’s uterus is precisely controlled by these transposons. Researchers have also observed similar control elements derived from viruses in humans and seven other mammalian species. It is believed that these ancient viral transpositions have been “domesticated” in the genomes long before the rise of modern viruses, serving an important role in the development of all mammalian embryos.
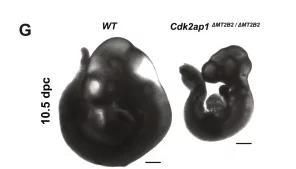
Figure: When transposons is eliminated, there is a delay in the development of mouse embryos. In the same lineage, transposons mutant (Cdk2ap1ΔMT2B2/ ΔMT2B2) exhibits a smaller size compared to the normal embryo (WT or Wild Type). This information is based on the original research paper.
The genomes of mice and humans share 99% similarity in protein-coding genes—they are highly similar. So where does the difference lie between humans and mice? One significant difference is in the control of genes. Even though humans and mice may have the same genes, they are regulated differently. Transposons, which help explain the differences between various species, play a major role in creating the diversity of gene regulation. This research suggests that the transposon of DNA elements derived from viruses is crucial for the survival of different species. When researchers removed these transposons from the genomes of mice, more than half of them died before birth.
According to another scientist involved in this research, Ting Wu, a professor at the School of Medicine at Washington University, this study reveals the potential impact of unforeseen alterations. Previously, transposons was largely considered redundant genetic material, yet it comprises a substantial portion of mammalian genomes. Numerous studies suggest that transposons plays a crucial role in shaping the human genome. Nevertheless, based on our findings, which, to the best of my knowledge, represent the first of their kind, we have demonstrated that the elimination of a segment of junk DNA can result in fatal characteristics, highlighting the indispensability of specific transposons.
It is worth mentioning that nearly 50% of human pregnancies are not successfully carried to term, and genetic causes are often not found. With this research, postdoctoral fellow Andrew Modjeski believes that exploring the relationship between transposons and pregnancy loss may provide some insights.
Origin of Species: satellite DNA variants
The second-mentioned research is not solely based on a single research paper. A group of researchers from the Whitehead Institute at the Massachusetts Institute of Technology has published several consecutive research papers on this topic for several years. They research satellite DNA. Their cumulative research suggests that satellite DNA is not junk or useless DNA but plays a crucial role in organizing the chromosomes within the cell’s nucleus.
I jokingly refer to satellite DNA as “parrot DNA”, because, like repeating learned words in a pet-parrot, satellite DNA consists of repeated sequences that tend to replicate. When the team at the Whitehead Institute began researching satellite DNA about seven to eight years ago, they had no particular interest in its variation. However, science often works so that unexpected patterns can be discovered even though research starts without prior assumptions. In a recently published research paper, the research team proposed that satellite DNA might contribute to the emergence of new species through speciation.
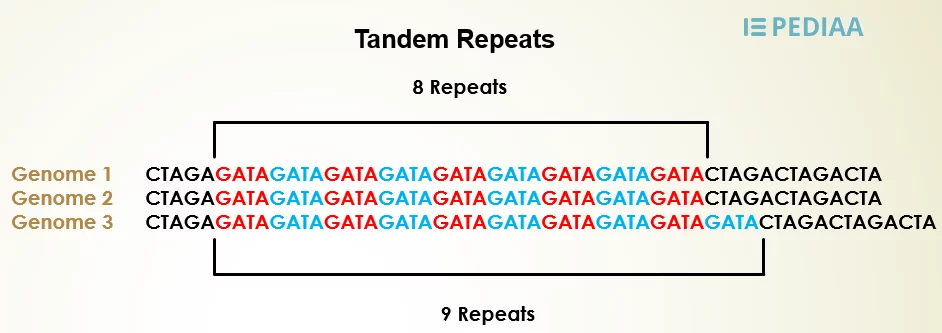
Figure: Satellite DNA is repetitive and tandemly repeated sequences of DNA.
Satellite DNA varies in quantity among different species. Even though protein-coding genes have a high degree of similarity (98-99%), junk DNA can exhibit significant differences. The most rapidly evolving portion of the genome is “junk” DNA. However, until recently, scientists wondered, “Since these are junk, why do they exist separately in different organisms?” Advancements in research and the Yama-Mashi lab have suggested that these repetitive “junk” DNA sequences may play a crucial role in the emergence of new species (speciation).
In the case of the fruit fly Drosophila melanogaster, there is a specific satellite DNA sequence called Prod. When scientists removed this Prod protein, they observed that the chromosomal nucleosome outside the nucleus of the fly became disorganized and formed a structure similar to a micronucleus. Furthermore, it was discovered that the flies died as a result. Surprisingly, researchers found that this particular satellite DNA sequence was absent in the closest relative species of Drosophila melanogaster. This observation provided them with the first clue to the mystery.
Suppose a small portion of satellite DNA is essential for the survival of one species of flies but absent in another species. In that case, it suggests that these two species have undergone distinct changes in their satellite DNA sequences for the same purpose. This raised the suspicion among Yama-Mashi and the research team that the inability to reproduce between the two species might be due to the differences in their satellite DNA.
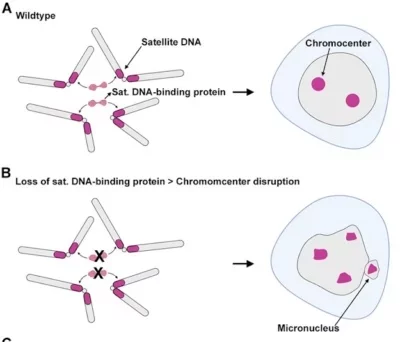
Figure: Satellite DNA keeps the chromosomes of normal flies together. However, in its absence, the chromosomes become disorganized and form a micronucleus structure. Image source: Original research paper.
To understand the correct interpretation of the variation in satellite DNA between two species of flies, the researchers initially relied on the phylogenetic tree of the fruit fly’s (Drosophila melanogaster) close relatives. They identified two closely related fruit fly species, Drosophila melanogaster and Drosophila simulans, which had diverged two to three million years ago.
These two species can reproduce artificially, but the offspring generally die or become reproductively sterile. When they examined the chromosomes of the hybrid offspring of these two species, they observed that the phenotype was strikingly similar to the satellite sequence-depleted chromosomes found in the fruit fly’s cell. The chromosomes were also fragmented outside the nucleus.
Interestingly, the researchers successfully created viable hybrid offspring of the two fly species by inducing specific mutations in the satellite DNA of the parental flies. These specific genes were named Hybrid Incompatibility Genes (HIGs). It was found that these genes were located within the satellite DNA itself. Further research revealed that these genes were associated with packaging the chromosomes of hybrids.
Their research showed that rapid mutations occur in satellite DNA, and to maintain the organization of chromosomes within satellite DNA, the associated proteins also need to undergo rapid changes. As a result, both species have developed their own “tricks” to work with satellite DNA. When the strategies for the same task differ between the two species, conflict may result in chromosomes being expelled from the nucleus.
Does junk DNA make the human brain unique?
Chimpanzees are humans’ closest relatives regarding evolutionary direction. Humans and chimpanzees share 98.8% of their DNA. Despite this high degree of similarity, there are differences in the brains of humans and chimpanzees. Our brain is about seven times larger than a chimpanzee’s, considering the size relative to the body.
The third research was conducted in Sweden. A group of researchers at Lund University investigated whether DNA separates the human brain from the chimpanzee brain. The answer lies in the non-genic region of the DNA.
However, they don’t work with giant humans or chimpanzees. Instead, they focus on stem cells. Stem cells are special types that give rise to all other specialized cells in the body. These stem cells are primarily reprogrammed from skin cells and then developed into brain cells, subsequently studied. Leading this research team is Professor Jacobson. He and his colleagues create brain cells from stem cells derived from humans and chimpanzees and begin to explore the differences between them.
It turns out that humans and chimpanzees use a specific portion of their DNA through separate processes. This portion plays a crucial role in brain development. This part is the structural variant of DNA, which is significantly longer and more repetitive than the primary DNA. It was previously referred to as “junk DNA” and was thought to have no function.
According to this research, it is understood that the formation and genetic processes of the human brain are much more complex than previously believed, with 98% of it being controlled by “junk” DNA.
Conclusion
The present time is very exciting for genomic biology. Through new sequencing technologies, it is now possible to easily create complete genomes through long-read sequencing. Furthermore, comparing multiple genomes has become much less challenging and expensive. With the advancement of these technologies, we can now ask questions about genomic biology that were previously impossible. Recent research in genomic biology has led to the discovery of the potential role of so-called “junk DNA.” A complete picture will be obtained with further evidence from future research.”
References:
- Modzelewski, Andrew J., et al. “A mouse-specific retrotransposon drives a conserved Cdk2ap1 isoform essential for development.” Cell (2021).
- Jagannathan, Madhav, and Yukiko M. Yamashita. “Defective satellite DNA clustering into chromocenters underlies hybrid incompatibility in Drosophila.” Molecular Biology and Evolution (2021).
- Johansson, Pia A., et al. “A cis-acting structural variation at the ZNF558 locus controls a gene regulatory network in human brain development.” Cell Stem Cell (2021).


Leave a Reply