Tag: Bioinformatics
-

Variant Calling for Microbial Genomes Using Short- and Long-Reads
Variant calling is a fundamental analysis in microbial genomics. It has a range of applications: diagnosing clinical pathogenic strains during an infection, understanding an epidemic, genotyping, and, more generally, studying evolution. There are so many SNP analysis pipelines. Then why am I writing this tutorial—and what new approach am I adding? Well, the pipeline in…
-

Course on Comprehensive Bioinformatics for Advanced Microbial Genomics
Oxford BioDiscovery এর সাথে Comprehensive Bioinformatics for Advanced Microbial Genomics কোর্সটির দ্বিতীয় ব্যাচ October 10, 2025 থেকে শুরু হবে। বাংলাদেশের বায়োকেমিস্ট্রি/বায়োটেকনোলজি/মাইক্রোবায়োলজি/জুলজি/বোটানি ইত্যাদি বিভিন্ন ব্যাকগ্রাউন্ডে থেকে এসে যারা মাইক্রোবিয়াল জিনোমিক্স নিয়ে NGS বিষয়ক গবেষণা করতে চান, তাদের জন্য এটি কম্প্রিহেনসিভ কোর্স হিসেবে ডিজাইন করেছি। গত মার্চে এটার প্রথম ব্যাচ শুরু করি, যা এই সামারে শেষ হয়।…
-
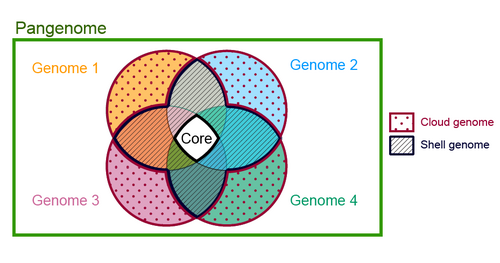
Microbial pan-genome analysis
Pan-genome means collection of ALL genome for a particular species. This is a fairly common analysis in microbial genomics. Compared to eukaryotic organisms, bacterial genomes a fluid. Take randomly two genomes from a bacterial species, chances are that there will be huge genetic differences in terms of gene content. That’s because, conjugation, recombination and horizontal…
-

Microbial ortholog gene clustering using real-dataset
Once we assembled and annotated bacterial genomes, next step after inferring phylogeny usually is to finding and clustering orthologous genes. What are orthologs? Well, Genetics 101 tells us that they are genes evolutionary shared between multiple taxa descended from common ancestors. That means a series of (pretty-much) reliable replications and cell division passed between these…
-

Deciding how to construct a phylogenetic tree
For any microbiology project, constructing a phylogenetic tree is an important analysis. A confusing one too — because there are many decisions to make along the way. These decisions can be both technical and hypothesis-driven. How to Build a Dataset for Phylogeny Let’s say someone has isolated and sequenced a few microbial strains. The most…
-

Annotation of Bacterial Genomes
This is Part 5 of tutorial series: NGS Workflow for Genome Assembly to Annotation for Hybrid Bacterial Data We’ll be use hybrid sequencing data (Illumina and Nanopore). This tutorial has five parts. Disclaimer: This post is a work in progress. This is genome assembly and annotation workflow that I use for microbial genomics. Previously, I used…
-

Hybrid Assembly of Bacterial Genome
This is Part 4 of tutorial series: NGS Workflow for Genome Assembly to Annotation for Hybrid Bacterial Data We’ll be use hybrid sequencing data (Illumina and Nanopore). This tutorial has five parts. Disclaimer: This post is a work in progress. This is genome assembly and annotation workflow that I use for microbial genomics. Previously, I used…
-

Microbial De Novo Genome Assembly from Long-Read Data
This is Part 3 of tutorial series: NGS Workflow for Genome Assembly to Annotation for Hybrid Bacterial Data We’ll be use hybrid sequencing data (Illumina and Nanopore). This tutorial has five parts. Disclaimer: This post is a work in progress. This is genome assembly and annotation workflow that I use for microbial genomics. Previously, I used…
-

Microbial De Novo Genome Assembly from Short-Read Data
This is yet another tutorial on microbial genome assembly. This is actually the Part 2 of tutorial series: NGS Workflow for Genome Assembly to Annotation for Hybrid Bacterial Data In this series we’ll be use hybrid sequencing data (Illumina and Nanopore). This tutorial has five parts. Part 1: Downloading and preparing data Part 2: Assembly with…
